Introduction
This tutorial is currently under development. However, here you can find diffusion MRI data acquired on a unique 300mT/m system (the Siemens Connectom system at the Massachusetts General Hospital). For more information please see the article mentioned below.
Diffusion MRI Scanning
A single volunteer underwent 8 hours of acquisition to provide uniquely wide and dense sampling of the available space of pulsed-gradient spin-echo (PGSE) measurements. The experimental protocol contains 48 HARDI shells, each with a different b-value arising from a unique combination of:
- gradient strength |G| = {60, 100, 200, 300} mT/m;
- pulse width δ = {3, 8} ms;
- pulse duration Δ = {20, 40, 60, 80, 100, 120} ms;
The b-values thus range from 50 to ~46,000 s/mm2, with echo times in the range 49 to 152 ms. The protocol is designed to cover the measurement space as widely as possible. The raw images, consisting of 4mm sagittal slices, have voxels with in-plane dimensions of 2mm x 2mm. The set of "blip-up, blip-down" directions in each HARDI shell comes from a 45-direction evenly-distributed point set taken from Camino and acquired in opposite directions to allow for eddy current distortion correction. The data was preprocessed with FSL using FNIRT and EDDY to correct for subject motion and eddy current distortions.
Data
For the time being we will provide the data as used in the ISBI 2015 white matter modelling challenge. We intend to release the remainder around the autumn of 2015. The challenge looked at a simple situation, two corpus callosum ROIs with 6-voxels each. One is from the genu, where the fibres are approximately straight and parallel, while the other is in the fornix, where the configuration of fibres is more complex.
The data and protocol are stored as gzip text files. The protocol's columns specify the gradient direction (columns 1 to 3), the gradient strength in T/m (column 4), Δ in seconds (column 5), δ in seconds (column 6), and the echo time TE also in seconds (column 7). The repetition time is 1 second. The image attached below details which data shells are included (in black) and which are not (in red).
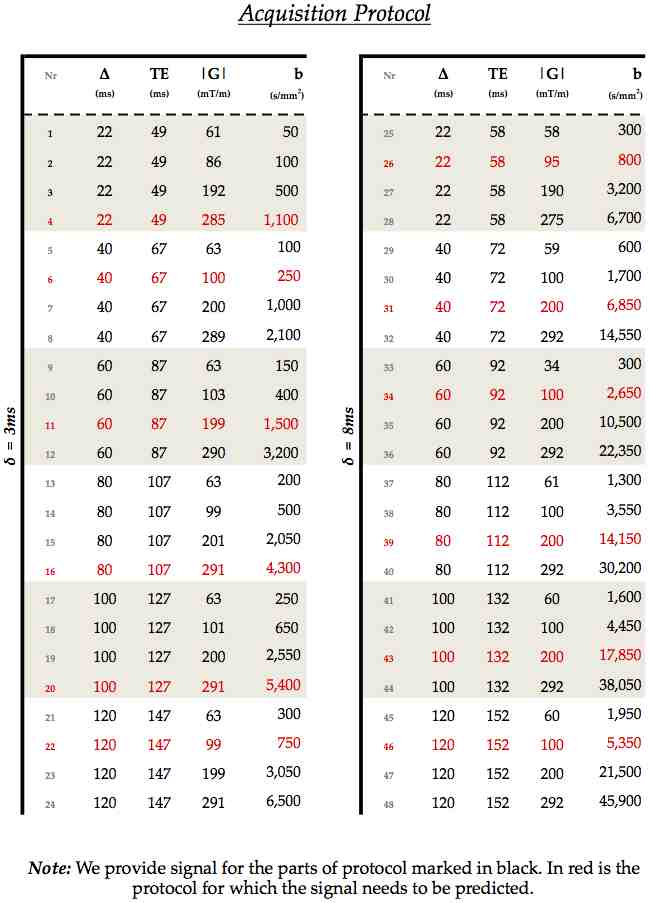
The genu and fornix signal datasets contains 36 shells, or 3,612 diffusion and non-diffusion weighted measurements: each shell has 90 diffusion-weighted and 10 interleaved b=0 measurements in each shell. (NOTE that you will actually find 12 other extra b=0 signals. This is because every one of the TE-specific sets had 401 measurements originally: one extra b=0 measurement preceded every 400=4x(90DW+10B0) acquisitions. Because we kept out 100 measurements for assessment, there is thus an extra b=0 preceding every 300 measurements instead.)
Using the data
If using this data in any publication, please cite the following paper: Uran Ferizi, Torben Schneider, Thomas Witzel, Lawrence L. Wald, Hui Zhang, Claudia A.M. Wheeler-Kingshott, Daniel C. Alexander, 2015. White matter compartment models for in vivo diffusion MRI at 300 mT/m. NeuroImage 118, 468-483.

